get_clades returns the tips that descend from a given node or time
depth on a dated phylogenetic tree.
get_clades(tree, cut = NULL, k = NULL)Arguments
- tree
is a dated phylogenetic tree with branch lengths stored as a phylo object (as in the
apepackage).- cut
the slice time
- k
number of slices
Value
A list of descendants
References
Schliep, K.P. (2010) phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593.
Examples
require(ape)
data(bird.orders)
plot(bird.orders)
axisPhylo(side = 1)
abline(v=28-23) # the root is here at 28
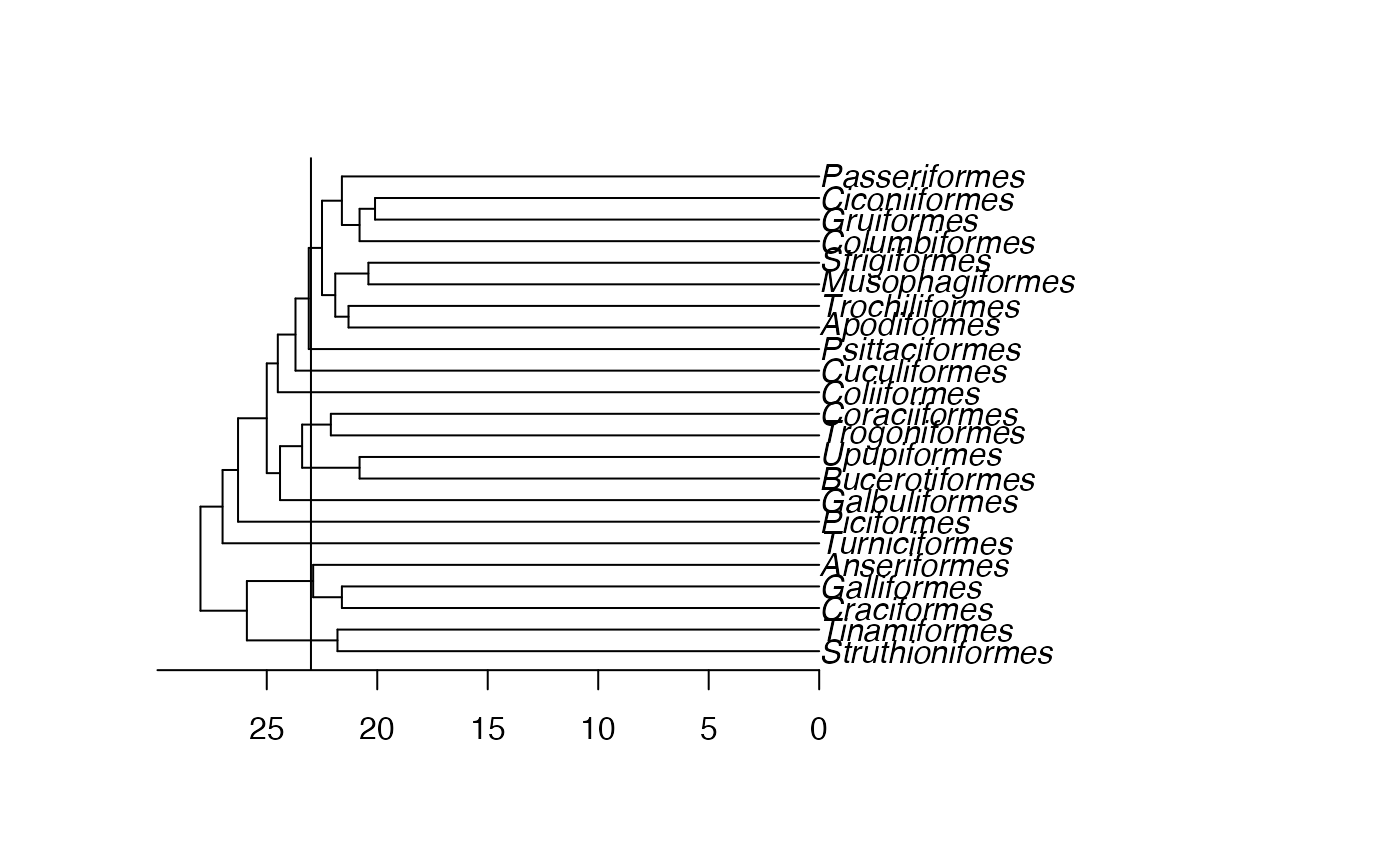 get_clades(bird.orders, 23)
#> [[1]]
#> [1] "Struthioniformes" "Tinamiformes"
#>
#> [[2]]
#> [1] "Craciformes" "Galliformes" "Anseriformes"
#>
#> [[3]]
#> [1] "Turniciformes"
#>
#> [[4]]
#> [1] "Piciformes"
#>
#> [[5]]
#> [1] "Galbuliformes"
#>
#> [[6]]
#> [1] "Bucerotiformes" "Upupiformes"
#>
#> [[7]]
#> [1] "Trogoniformes" "Coraciiformes"
#>
#> [[8]]
#> [1] "Coliiformes"
#>
#> [[9]]
#> [1] "Cuculiformes"
#>
#> [[10]]
#> [1] "Psittaciformes"
#>
#> [[11]]
#> [1] "Apodiformes" "Trochiliformes" "Musophagiformes" "Strigiformes"
#> [5] "Columbiformes" "Gruiformes" "Ciconiiformes" "Passeriformes"
#>
get_clades(bird.orders, 23)
#> [[1]]
#> [1] "Struthioniformes" "Tinamiformes"
#>
#> [[2]]
#> [1] "Craciformes" "Galliformes" "Anseriformes"
#>
#> [[3]]
#> [1] "Turniciformes"
#>
#> [[4]]
#> [1] "Piciformes"
#>
#> [[5]]
#> [1] "Galbuliformes"
#>
#> [[6]]
#> [1] "Bucerotiformes" "Upupiformes"
#>
#> [[7]]
#> [1] "Trogoniformes" "Coraciiformes"
#>
#> [[8]]
#> [1] "Coliiformes"
#>
#> [[9]]
#> [1] "Cuculiformes"
#>
#> [[10]]
#> [1] "Psittaciformes"
#>
#> [[11]]
#> [1] "Apodiformes" "Trochiliformes" "Musophagiformes" "Strigiformes"
#> [5] "Columbiformes" "Gruiformes" "Ciconiiformes" "Passeriformes"
#>